Research
We work on Human disorders. Some of our research is on race-specific alterations, machine learning, and microbiota, breast cancer, PTSD, pharmacophores. In Chitrala lab, we focus on resolving some of the problems in healthcare, and here are some of our recent findings.
Here are some research details:
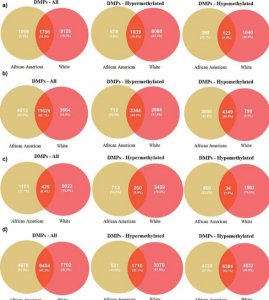
- Race-specific alterations in DNA methylation among middle-aged African Americans and Whites with metabolic syndrome. Metabolic syndrome (MetS) is a cluster of cardiometabolic risk factors for all-cause mortality, cardiovascular disease, and cancer. We focus on identifying epigenetic alterations associated with MetS in African Americans and Whites thereby providing insight into the genes influencing different health outcomes.



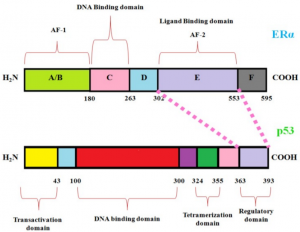
- Analysis of the TP53 Deleterious Single Nucleotide Polymorphisms Impact on Estrogen Receptor Alpha p53 Interaction: A Machine Learning Approach. In this research we focus on how estrogen plays a key role in breast cancer and how these mutations impact p53 protein product. We develop machine learning models to unravel the atoms, residues and solvent accessibility surface area impacting this interaction. These findings will be crucial in decision making for hormone-based therapies targeting breast cancer.



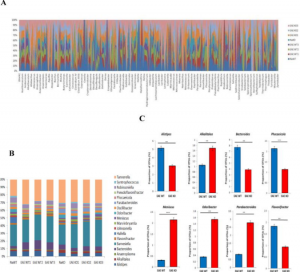
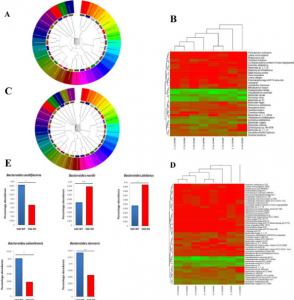
- CD44 deletion leading to attenuation of experimental autoimmune encephalomyelitis results from alterations in gut microbiome in mice. CD44 plays a crucial role in the regulation of experimental autoimmune encephalomyelitis a mouse model of Multiple sclerosis. We focus on how knocking of CD44 gene in mouse models result in the alterations of gut microbiota and short-chain-fatty acids. We aim to study on how gene phenotype of knock-out animals shape the gut microbiome.

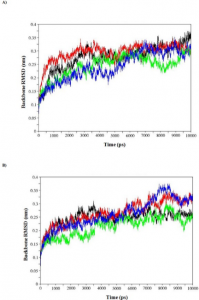
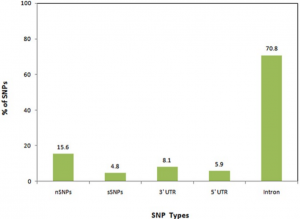
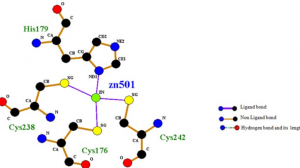
- Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 gene. TP53 is the most mutated gene in breast cancer and is a major genetic risk factor. TP53 cause chemo resistance and has a poor prognosis. Our study focus on how deleterious non-synonymous single nucleotide polymorphisms in TP53 impact the p53 protein product.

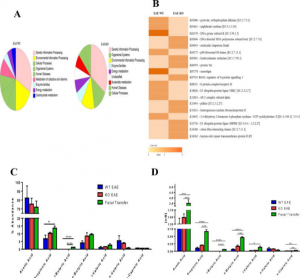
- Prediction of Possible Biomarkers and Novel Pathways Conferring Risk to Post-Traumatic Stress Disorder. We focus on elucidating the novel biomarkers and pathways conferring risk to PTSD. These identified biomarkers could be key targets for PTSD.

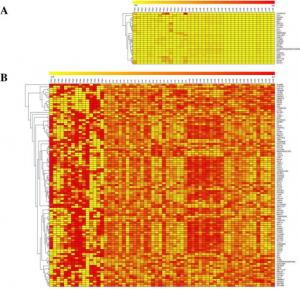
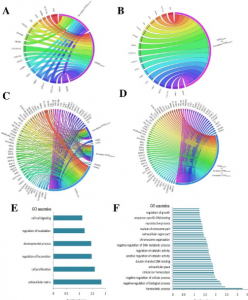

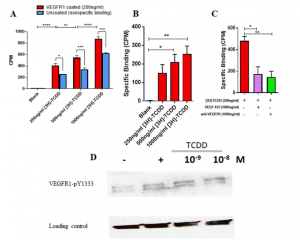
- Computational prediction and in vitro validation of VEGFR1 as a novel protein target for 2,3,7,8-tetrachlorodibenzo-p-dioxin. We focus on identifying the novel protein receptor targets for TCDD using computational and in vitro validation experiments.